What mechanisms drive pervasive regulation of translation and protein degradation in meiosis? And what can this teach us about cellular aging?
Degradation
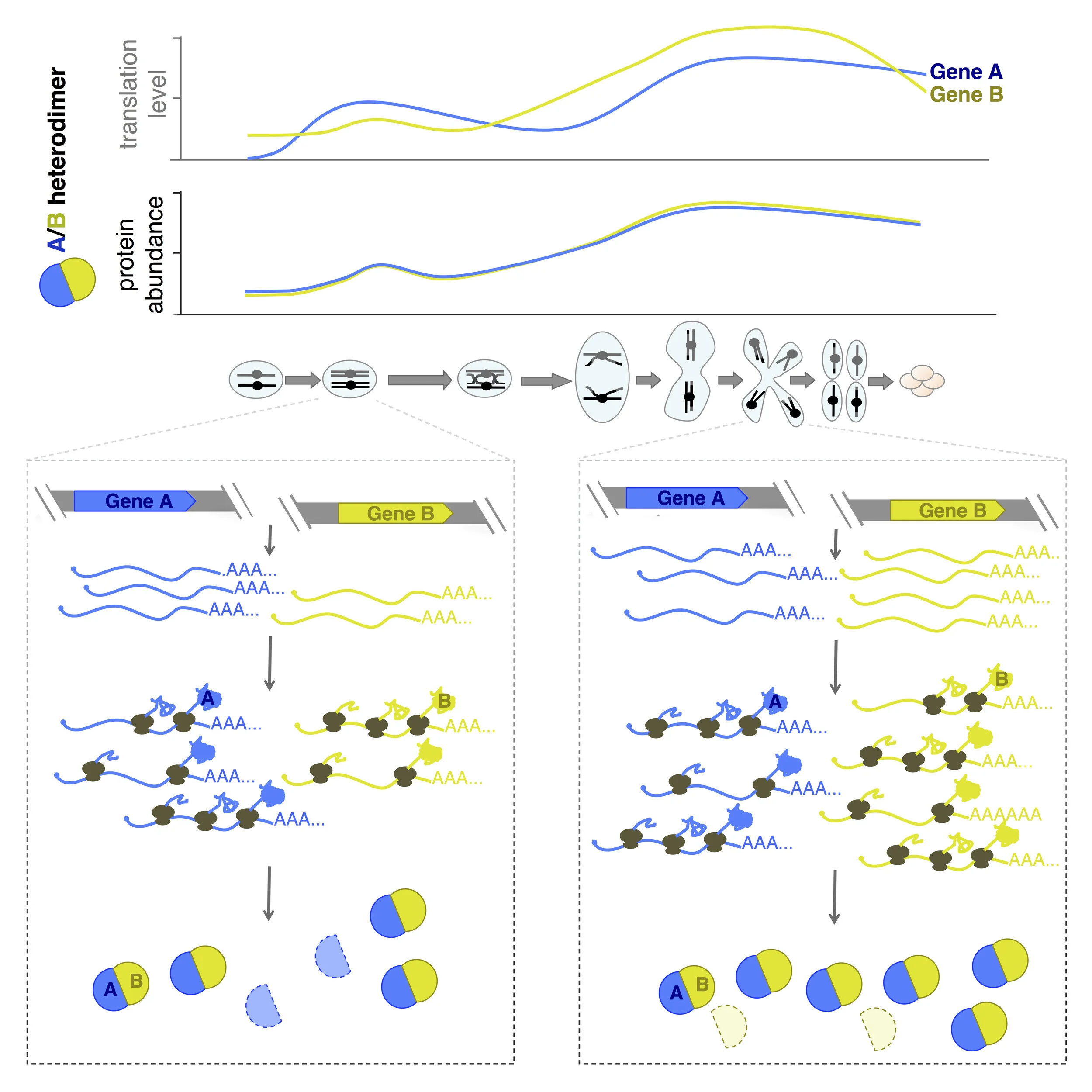
Our study of parallel mRNA, translation, and protein measurements during meiotic differentiation excluded analysis of protein degradation, which we could not measure directly because of the recalcitrance of meiotic yeast cells to exogenous amino acid labeling approaches. Because protein dilution (the major cause of protein level down-regulation in mitotic cells) is not seen during meiosis in yeast, however, decreases in simple protein levels measured over time appeared to be indicative of protein degradation. This conclusion was based on analyses of well-studied meiotic degradation targets and classical confirmatory approaches on cases of interest. Our analyses suggested that essentially all proteins are actively degraded at some point in the meiotic program and comparison of protein patterns over time by hierarchical cluster analysis could be used to predict groups of proteins that were degraded by the same mechanism. Our most intriguing observation, however, came from comparison of translation and protein patterns over time for members of heterodimers or larger protein complexes. Among a group of meiotic genes involved in homologous recombination and synaptonemal complex formation that are co-regulated at the level of translation, patterns of protein over time diverged, except for those that physically interacted, which even more closely matched each other than patterns of translation for these same genes. Because the post-translational matching among this group of genes was specific to proteins that stably physically interacted in complexes, these results suggested that non-interacting protein partners specifically were degraded as a natural part of meiosis.
Protein-level matching of physically interacting proteins over time. Top: translation levels over time and protein levels over time are shown for a typical heterodimer during meiosis, revealing high matching specifically at the protein level. Below: our interpretation of the regulation responsible for these patterns, involving protein degradation of uncomplexed partners.
We found that protein-level patterns over time for proteins that constitutively physically interact matched each other much more closely than translation-level patterns for these same proteins as a general rule in meiosis . In contrast, groups of genes that were co-regulated in synthesis of their protein partners, for example sequential enzymes in linear biosynthetic pathways, had a similar degree of translation-level matching as physically interacting protein partners, but much lower protein-level matching. This result was important because it revealed that the apparent matching of protein levels for interacting proteins was biological and not a feature of the difference in measurement properties for translation, which are instantaneous and sequencing-based, and protein, which are mass-spectrometry-based but not pulse-labeled. Interestingly, in a few cases, heterodimer partners were translated (and had protein levels) with precisely matching patterns over time. This was exemplified by TUB1/TUB2, the dominant tubulin heterodimer, which is encoded by two genes with dissimilar promoters and UTRs. The TUB1mRNA is subject to splicing, whereas TUB2 is not, and yet their synthesis patterns are indistinguishable, suggesting that eukaryotic cells have the ability to precisely coordinate synthesis, although this is not the norm in most cases.
We believe that our study represents the first such case, and supports a model of programmed “sloppy” synthesis matching of protein partners followed by post-translational degradation of uncomplexed partners as the rule during meiotic differentiation, despite the ability of meiotic cells to perfectly match synthesis in a few specific cases. The two exceptional classes of complexes in this study are of particular interest to us and are being investigated more deeply First, we would like to understand the reason that meiotic cells effectively perfectly match synthesis in a few cases, like TUB1/TUB2. The fact that excess Tub2 is known to be toxic suggests an evolutionary advantage to the additional synthesis regulation in this case and we are testing whether this explains the other cases of this type, as well. The other exceptional class of protein complexes of interest include cases in which most but not all members of a complex are precisely matched at the protein level. We noted that the unmatched components tended to serve regulatory roles and believe that this pattern is diagnostic of the presence of a “linchpin” mechanism by which a single protein can control the activity of a whole complex. The regulation and roles of most well characterized protein complexes are unexplored in meiosis. We believe that our dataset and analytical strategy gives us a foothold into identifying meiotic roles for protein complexes with this signature, which include many important “housekeeping complexes”.
Protein Degradation and Aging
A hallmark of aged cells is the breakdown of protein integrity, or proteostasis, which results from the damage to key proteins and complexes over time, leading to more accumulated damage, and ultimately cell dysfunction. The specific proteome components that are most susceptible to damage and that drive its accumulation remain unclear, but the survival of future generations depends on protection of one cell type—gametes—from inheriting damaged components from their precursor cell. As the sole components that create the next generation, their fitness is critical. During gametogenesis in the simple budding yeast, as a precursor cell is differentiated into gametes, we observe the degradation of many cellular structures and proteins, followed by their resynthesis and reorganization. This cellular restructuring is associated with an active rejuvenation program that allows equivalently young gametes to be produced from old or young precursor cells. The mechanisms that contribute to this natural rejuvenation program are not known, but the Ünal lab has shown that it can be recapitulated by exogenous expression of a meiotic transcription factor in aged mitotic cells, suggesting that it is portable.
Gametogenesis/meiosis in yeast thus offers the opportunity to watch as the cell shows us what proteins and complexes it needs to reset and reorganize to ensure cellular youth, and the mechanisms it uses to achieve this. Of particular interest are proteins of basal or “housekeeping” function, which are long-lived in mitotic cells, but degraded and replaced at great energetic cost during mid- to late-gametogenesis, suggesting that yeast cells remodel their proteome during gametogenesis as a quality control measure. We are systematically identifying the key set of cellular components that are reset as gametes are created from precursor cells. This project will build an atlas to reveal the proteins and complexes that are important enough for young cell identity to warrant the energetic cost of resetting them during gamete formation, as well as those that may be toxic enough to warrant their active removal. Essentially, we aim to ask cells which proteins are valuable enough for cellular youth to warrant the high cost of their resetting during gametogenesis. Because only 10% of proteins are actively degraded in mitotic cycling cells but over 90% are actively degraded in meiosis, this project will also allow us to identify many new mechanisms of protein degradation.
The timing of A) Chromosomal events for meiosis; B) Other gametogenic remodeling events; C) Protein abundance for stages measured, with hours in gametogenesis above; D) Known degradation mechanisms. Timing is aligned vertically for all panels.
This project is a collaboration with Marko Jovanovic.
Pervasive Noncanoncial Translation
Our original study of new protein synthesis in meiosis showed that translational regulation contributes strongly to temporal tuning of gene expression through meiosis, with 24% of genes subject to strong translational control during meiotic progression. This dataset provides a valuable tool to probe the molecular basis for, and cellular importance of, such dynamic control and the identification of the cis- and trans-factors responsible. We are currently investigating several examples of such factors, including prominently the translation of upstream Open Reading Frames (uORFs).
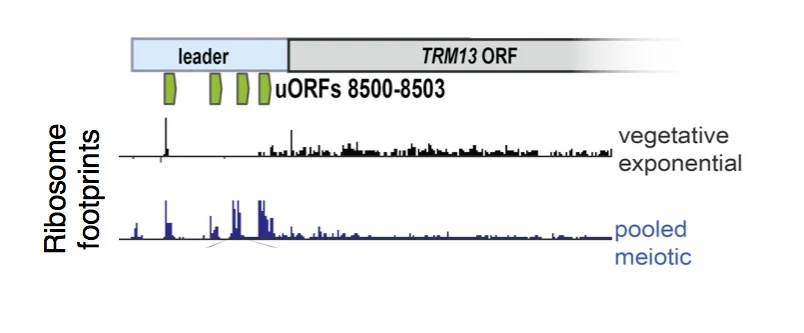
uORF translation is enriched in meiotic cells. Ribosome footprints are plotted over a sample gene, TRM13, showing enrichment for ribosomes in the 5' leader region (commonly called the 5'UTR) in meiosis. These uORFs (annotated in green) represent just 4 of over 10,000 uORFs that we identified to be translated in meiosis, suggesting a noncanonical mode of translation in these cells.
Thousands of instances of uORF translation were observed in cells undergoing meiosis, suggesting a meiotic shift in translation mechanism for at least a subset of transcripts. Additionally, many of these uORFs initiate translation at near-cognate (non-AUG) codons. The uORFs observed are highly discrete, specific, and heavily enriched in meiosis relative to any other condition examined to date, but the molecular basis for their translation remains a mystery. We aim to unravel both the biological significance of these uORFs as well as the translation specializations that produce them in a meiosis-specific manner. We recently found that low meiotic levels of eIF5A, a translation factor implicated in elongation and start codon selection, drive part of the increase in non-AUG initiation seen in meiotic cells, but believe that additional factors are involved, as well.
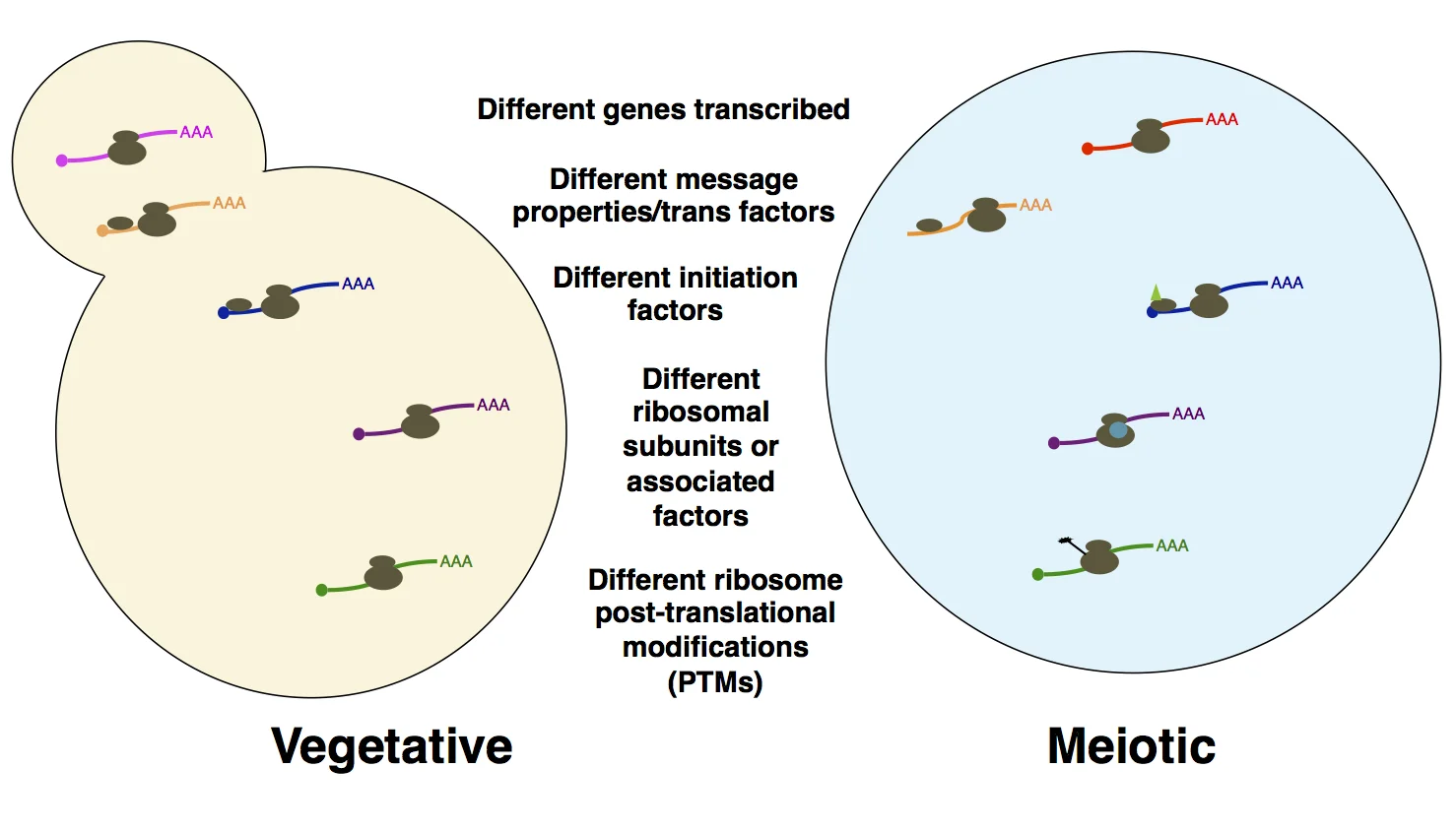
Probing the basis for noncanonical translation in meiotic cells. A number of factors that may be responsible for the unusual degree of dynamic translational regulation and uORF translation in meiosis are shown. We are systematically probing examples of cis- and trans- factors like those outlined here, with the goal of understanding what is special about translation (particularly translation initiation) in meiosis.